- Posted by admin
The Human Microbiome and I-Cubed: A Modern Medical Paradigm
The human microbiome, defined as the collection of microorganisms that reside within our body, have coevolved over the history of mankind, and have been overlooked as determinants of health and disease. Given the appearance of new microbial agents and the everyday occurrence of unexplained lethal neurological syndromes of suspected infectious etiology, scientists have begun to identify a plethora of microbial agents in our body and in the human genome. The true capability of such microbes residing in our body to cause human disease has become the focus of medical science. Post-infectious autoimmune illness is increasingly recognized due to resident and invasive microbial agents that have the capacity to trigger our immune system, turning it on and off at will. With differences in resident microbial niches, imperfect host defenses, and susceptibility to epidemic and endemic diseases in the environment, there are ever increasing opportunities for infectious bacterial, virus, and parasitic exposures.
The triad of infection, immunity and inflammation, abbreviated as I-Cubed I3posits that complex self-sustaining responses from infectious insults to the nascent immune system, whether by contagious or innate sources endemic to ones microbiome, can begin to explain the etiopathogenesis of diverse medical, neurologic and neuropsychiatric disorders. Underlying genetic vulnerability, pre- and post-natal factors, and individual differences in exposure timing and host immune responses, may further explain differences in eventual outcome. Innovative therapeutic strategies involving immune modulation may be effective in some affected patients.
THE HUMAN MICROBIOME
For an appreciation of the human microbiome, imagine the excitement of a scientist at a medical conference claiming to have discovered a new human organ which like the immune system, contains collections of cells and a 100-times more genes than the host. Not only is it tailored to the individual host, but it is modifiable by stress, diet, medications, exercise, and antibiotics. When lost, nearly all aspects of the host’s normal physiological function are altered. Although it has been known for some time that the human body is inhabited by resident flora in a factor greater than 10:1, most researchers have focused instead on a minority of disease-causing or “pathogenic” organisms with far fewer examining the benefits of the resident bacterial flora. The completion of the human genome sequence in 2001 which was the crowning achievement in biology, was incomplete because it did not look at the synergistic activities of humans and microbes living together. With many well recognized neurological diseases of likely infectious trigger yet unassigned to infectious microbes such as most cases of aseptic meningitis, encephalitis, and cerebral vasculitis, there was a need for a second human genome project to provide a comprehensive inventory of microbial genes and genomes at major sites of microbial colonization in the human body. Many investigators envisioned that understanding the microbial contributions to inflammatory disease could be addressed effectively through a thoughtful integration of modern technologies and clinical insight.
The concept of the human microbiome or microbiota originated with the Rockefeller University scientist Joshua Lederberg, as an ecological community of commensal, symbiotic, and pathogenic microorganisms sharing our body space. It is estimated that 20% to 60% of the human-associated microbiome, depending on body site, is still resistant to conventional culture techniques making it difficult to accurately estimate its true diversity. More recently, the human microbiome has been studied in different biological states using gene sequencing techniques. Scientists have used molecular tools to extract and compare bits of a particular kind of ribonucleic acid (RNA) the products of deoxyribonucleic acid (DNA) transcription and translation, to determine if previously known or new microbes were present in a particular human tissues such as blood. This technique which is widely used as a biomarker for microbial disease uses a particular kind of RNA known as 16S ribosomal RNA (rRNA). Since the genes for ribosomal RNA have changed little over millions of years as organisms have evolved, slight changes in their composition provide valuable clues to the very nature of microbial organisms located in the human body. The 16S rRNA gene is very short, just 1,542 nucleotide bases making it quickly and cheaply copied, sequenced and then compared to libraries of stored 16S rRNA genes from numerous known bacteria. The ones that match up perfectly are microbes that have been previously identified while others that show differences may be previously unknown microbes. Such studies of gastrointestinal microbes at the 16S rRNA gene level have revealed significant diversity in the flora of individuals. We are presently at a public health crossroads, in a position to make gigantic gains in our knowledge to better understand how microbes impact on human health, transitioning from description to causality and microbial engineering. Underscoring this, two papers published simultaneously in the journals Science and Nature called for the establishment of a Unified [domestic] Microbiome Initiative and International Microbiome Initiative.
With more than 90% of cells in the human microbiome understood to be bacterial, viral, parasitic, fungal or otherwise non-human in nature, and human metabolism and immunity attributed to the molecular genetic contribution of microbial and human interaction, human beings are being referred to as superorganisms. In the last decade the United States (U.S.) Human Microbiome Project and European MetaHit, two large-scale genomic projects, along with several private efforts, have investigated the microbiota in a variety of human body niches using new molecular genetic tools. While many sites such as the skin, oral and nasal cavities, and vagina are all relatively easy to access, the majority of research in this area has focused on the gastrointestinal tract, in particular the colon. A newly recognized axis of communication between the gut and brain have led to the recognition of a mind-gut connection attempts to explain the spectrum of functional symptoms from anxiety and depression to irritable bowel syndrome. With five phyla representing the majority of bacteria that comprise the gut microbiota, there are about 160 species in the large intestine alone of any individual, and very few of these are shared between individuals. The functions contributed by these species appear to be found in everybody's gastrointestinal tract, an observation that suggests that microbial function is more important than the identity of the species providing it. Our understanding of human microbial biology first derived from pure cultures and genomic sequencing, has been limited by sampling bias toward four bacterial phyla, Proteobacteria, Firmicutes, Actinobacteria and Bacteriodes, out of the 35 bacterial and 18 known archael phylum-level lineages. With roughly two-thirds of published microbiological research dedicated to only eight bacterial genera, all of which grow well on agar culture plates, it is unlikely that they are representative of the 5,000 or more species known to us.
I-CUBED: INFECTION ACTIVATES IMMUNITY
Immunity was historically understood for its purported effects due to active immunization. The first type was the effect of immunization that resulted in definable changes in the cell-free bodily fluid or serum or humor while another was the observed protective effect associated with multiplication of specific cells. Two primordial types of immune cells are now recognized, one lineage termed B-cells that mature in the bone marrow and further differentiate into plasma cells and memory cells. Mature plasma cells are capable of producing antibodies capable of latching onto their target in a lock-and-key specific fashion when their surface antibody receptors recognize other cells displaying foreign antigens. Other B-cells mature into memory B-cells that circulate in the blood stream. The other cell lineage, termed T-cells, also derived in the bone marrow, instead pass though the thymus gland where they achieve their final maturation, and are thought to be most protective in recognizing virus-infected cells. These cells participate in the defense against intracellular bacterial, fungi and protozoan infections; cancers and transplant rejection. Other aspects of enhanced cellular immunity include the secretion of cell-signaling molecules termed cytokines that promote cell-to-cell communication in immune responses and stimulate the movement of cells towards sites of inflammation and infection.
Not surprisingly, major understandings of the pathophysiology of autoimmune diseases has been achieved through an appreciation of infectious triggers of the humoral and cell-mediated immune system. When first identified as the causative agent of the neurological disorder, Whipple disease almost 25 years ago by Relman and coworkers, it was unclear whether the uncultured bacillus Tropheryma whippelii was a rare member of the normal human microbial flora and whether it might be associated with other human diseases. Whipple disease causes a systemic infection which unrecognized and therefore untreated, involves the gastrointestinal tract, heart, and brain. According to phylogenetic analysis the isolated bacterium was a gram-positive Actinomycetes not closely related to any known genus. A molecular genetic approach amplifying a 16S rRNA sequence directly from tissues of five unrelated patients determined its nucleotide sequence. A decade later, the same authors performed ultrastructural studies of intestinal biopsy specimens from affected patients. These studies showed the location of Tropheryma whippelii rRNA to be most prevalent near the tips of the intestinal villi in the lamina propia just basal to the epithelial cells, located between cells and not intracellular indicating that the bacillus grew outside cells and that it was not an obligate intracellular pathogen. Such studies ushered in a generation of molecular genetic technology used today in the study of resident human microbes.
Relman later observed that molecular, cultivation-independent methods revealed that the distribution and diversity of microorganisms in the world was far greater than previously appreciated. One particular molecular genetic technique compared human-tissue derived DNA sequences with those of known pathogenic and commensal bacterial, viral, fungal and protozoan genomes in established expressed-sequence tag libraries. However inefficient and cost-ineffective for screening large numbers of specimens in most laboratories, it revealed surprising findings of nonhuman genetic sequences that appeared to be an inherent feature of the human genome. It appears that all humans have human endogenous retrovirus sequences as an integral part of their genome. At some time during the course of human evolution, exogenous progenitors of the human endogenous retrovirus inserted itself into the human germ-line reproductive cells where they were replicated along with the host cellular genes. However intact disease-producing retroviruses differ in the presence of at least one additional coding region, the env gene that encodes viral membrane proteins that mediate the budding of virus particles to the cellular receptors enabling virus entry as the first step in the pathway to a new replication cycle and disease pathogenicity.
Hajjeh and coworkers observed that unexplained deaths and critical illness possibly due to infectious causes in previously healthy persons occurred at an incidence rate of 0.5 per 100,000 per year from 1995 to 1998 among 7.7 million persons in four U.S. Emergency Infectious Programs. However, only two-thirds of which were diagnosed by reference serological tests and the remaining one-third by PCR based methods. These findings suggested the need for molecular genetic surveillance approaches to detect present and emerging infectious diseases. New molecular biological techniques have led to the identification of several previously uncultivable infectious agents such as non-A and non-B hepatitis, and Hantavirus. Real-time PCR methods with primers and a probe targeting conserved regions of the bacterial 16S rRNA revealed rRNA in blood specimens from healthy individuals raising the possibility that there were normal populations of bacterial DNA sequences in the blood compartments previously been considered sterile at least most of the time. While persistent infection is potential source of nonhuman sequences in normally sterile human anatomic sites, not all bona fide pathogens have been associated with pathology.
The immunological mechanisms and interactions between resident microbial agents and the human host have been studied at various body sites. The interaction between resident oral bacteria and human gingival epithelial cells in culture demonstrate their potential for virulence. The microbial agents frequently associated with periodontal diseases, include Bacteroides forsythus, Campylobacter curvus, Eikenella corrodens, Fusobacterium nucleatum, Porphyromonas gingivalis, and Prevotella intermedia. The effects of these bacteria on the production of interleukin-8, a proinflammatory chemokine, were also measured. Fusobacterium nucleatum adheres to, and invades human gingival epithelial cells accompanied by high levels of interleukin-8 secretion from epithelial cells. By electron microscopy, this invasion occurs via a zipping mechanism that requires the active involvement of actins, microtubules, signal transduction, protein synthesis, and energy metabolism of the human gingival epithelial cells, as well as protein synthesis by Fusobacterium nucleatum.
Other investigators have noted a heightened risk of inflammatory bowel disease, spondyloarthropathy, and colorectal cancer between diffusely adherent Eschericia coli and areas of dysplastic mucosa of the colon that made it easier for the bacterial pathogens to gain direct contact with the mucosal surface, a location that is relatively sterile in the normal colon. This allowed interaction between bacterial components and intrinsic T-cell receptors of the human mucosa with subsequent downstream protein signaling as the mechanism for early oncogenesis illustrating yet another molecular genetic property of the resident bacteria in their putative role in genotoxicity and human disease. If epithelial-associated bacteria play a causative role in inflammatory bowel disease and colorectal cancer, then dietary consumption of soluble plant fibers that prevent mucosal recruitment of bacteria may be protective against both conditions.
Post-infectious autoimmunity is a recognized phenomenon with several theories to explain its occurrence including molecular mimicry, bystander activation, and viral persistence. Alone or in combination, these mechanisms have been used to account for the immunopathology observed at the site of infection and in distant areas of the body. Molecular mimicry occurs when there are shared immunologic identities or epitopes between the microbe and host. One well recognized example is rheumatic fever, a systemic autoimmune disease that occurs after group A beta-hemolytic strep infection wherein affected patients develop and manifest circulating reactive antibodies to the bacterial organism reactive to the heart, joint, and brain leading to the cardinal manifestations of rheumatic fever. Pediatric autoimmune neuropsychiatric disease associated with GABHS infection or PANDAS, is another example of bacterial-based molecular mimicry. There is a reciprocal dynamic between the central nervous system and immune system, conditioned by the microbiome that regulates a cross-talk which is easily perturbed by pre- and post-natal exposure to pathogens and genetic vulnerability toward dysimmunity, the timing of which may be important determinants of diverse neuropsychiatric illnesses.
Viruses with cross-reactive epitopes to hepatitis B virus and myelin basic protein, a constituent of myelin, develop autoimmune experimental allergic encephalomyelitis due to circulating T-cells that preserve the memory of the virus and cross-react with myelin present in brain white matter of experimental mice. There is a form of post-infectious encephalitis named acute disseminated encephalomyelitis, an inflammatory demyelinating disorder of the brain in children that follows seemingly minor viral infection with a 2 to 30 days latency period that is thought to be post-infectious and autoimmune. It is believed that naissance of autoimmunity in such disorders originates when novel disease-inducing autoantigens are presented by specialized elements of the immune system in a tri-molecular complex comprised of antigen-presenting cells, major histocompatibility complex class II molecules and autoreactive CD4+ T-cells.
Bystander activation and killing, a second mechanism that can also lead to autoimmune disease, has gained support through the use of experimental animal models mirroring some of the features of autoimmune disease such as the non-obese diabetic mouse for type 1 diabetes and experimental autoimmune encephalitis. It states that virus infections lead to significant activation of antigen presenting cells that potentially activate pre-primed autoreactive virus-specific T-cells that migrate to areas of virus infection/antigen such as the pancreas or brain. There, they encounter virus-infected cells presenting certain molecular tags, in turn releasing cytotoxic granules resulting in the killing or death of the infected cells. The dying cells, CD8+ T-cells and inflammatory cells within such inflammatory foci release cytokines that lead to the demise of uninfected neighboring cells and additional immunopathology at sites of infection.
Persistent viral infection is a third mechanism of immune-mediated injury due to the constant presence of viral antigens that in turn driving the immune response. Yet unproven in humans, an example of this occurs in experimental mice who develop a condition termed Theiler murine encephalomyelitis in which persistent infection leads to a T-cell-mediated immunopathology in genetically susceptible animals. Susceptible strains develop virus-specific delayed type hypersensitivity responses while resistant strains do not. This response has been proposed as the basis for flaccid paralysis that spread rapidly to all four limbs after an incubation period of 7 to 30 days due to inflammation and demyelination in the brain and spinal cord.
I-CUBED IMPLICATIONS FOR THE BRAIN
The implications of post-infectious mechanism for the brain are boundless and far-reaching for understanding autoimmune encephalopathy. Once considered a black box, the blood-brain barrier is becoming better understood for its constituent composition of capillary endothelial cells joined at several points by tight junctions (figure 1 and 2). When disrupted, it allows the transudation of cellular elements and inflammatory cytokines and other inflammatory intermediates from the systemic circulation to enter the unprotected brain, therein promoting a chain reaction of local inflammatory changes that expresses itself as encephalopathy or a mind or thinking disturbance. The availability of new brain imaging techniques able to sample the function of the blood-brain barrier (single photon emission computed tomography [SPECT]) and intrinsic metabolism (positron emission tomography [PET]) has begun the clarify the “supply and demand” equation of the brain function (figure 3 and 4).
CONCLUSION
Medical science is just now realizing the full importance of the microbial world. Thanks to genetic advances such as low-cost high-throughput sequencing of microbial communities comprising the human microbiome, the identity and function of uncultivable microbes is being unveiled. Beyond improved microbial cataloguing, we are learning that human beings are superorganisms integrating the identity, function and immunity of resident bacterial, yet prepared throughout own innate and adaptive immune systems, to deal with invading organisms. Modified by genetics and environmental factors, factors, once protective immunity has become the source of autoimmunity through a variety of well-established mechanisms. Public health officials and neuroepidemiology researchers will be called upon to guide the understanding of I-Cubed illnesses and the implications of the human microbiome for communicable and non-communicable diseases, at times one leading to the other, as the natural history is appreciated and the responsiveness of given medical and neurological disorder to a variety of medical approaches including strong antibiotics and immune-modulatory is established.
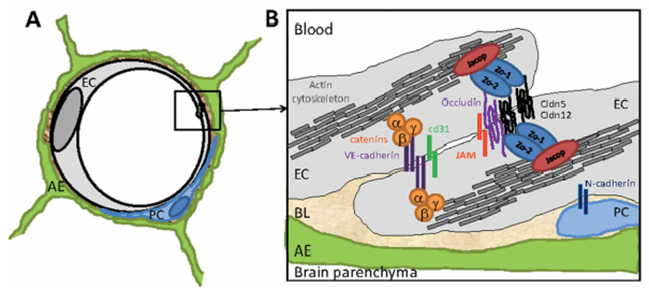
1. A. Cross-section schematic representation of a capillary in the human blood-brain barrier over an endothelial tight junction. B. The insert shows the molecular composition of tight and adherens junctions. See text for details. Reproduced from, Daneman R. The blood-brain barrier in health and disease. Ann Neurol 2012; 72:649, with permission of the publisher.
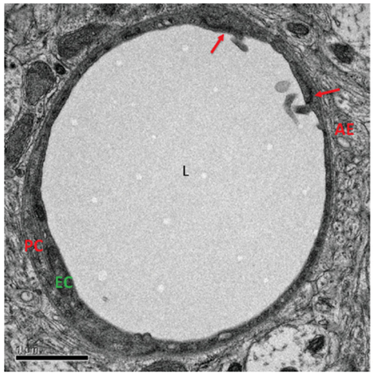
2. Electron micrograph of a capillary in the adult murine blood-brain barrier. Endothelial cells are held together by tight junctions (red arrow). Reproduced from, Daneman R. The blood-brain barrier in health and disease. Ann Neurol 2012; 72:650, with permission of the publisher.
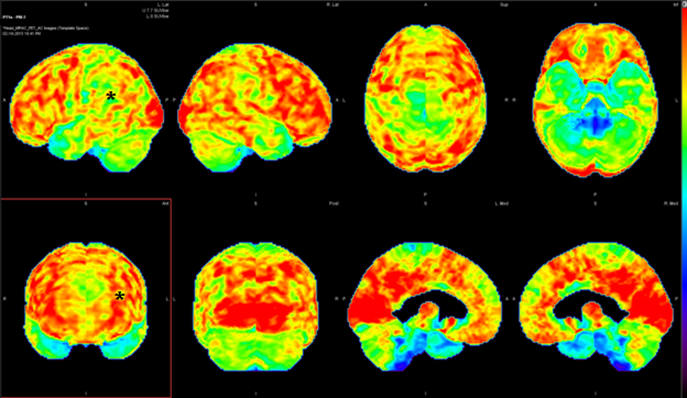
3. Magnetic resonance imaging (MRI) combined with Fluorodeoxyglucose positron emission tomography (FDG-PET). A 12-year-old girl with post-infectious autoimmune encephalopathy. There is hypometabolism of the left temporoparietal region (*).
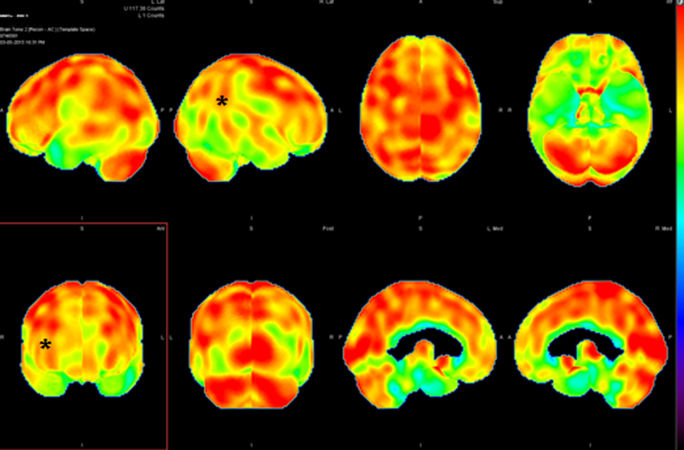
4. Nuclear medicine cerebral perfusion (brain) single photon emission tomography (SPECT). A 17-year-old girl with post-infectious autoimmune encephalopathy. There is hypoperfusion of the right temporoparietal region (*).
REFERENCES
Regidor E, de la Fuente L, Gutierrez-Fisac JL, et al. The role of the public health official in communicating public health information. Am J Public Health 2007; 97:S93-S97.
Lederberg J, McCray AT. 2001. ’Ome Sweet ’Omics—a genealogical treasury of words. New Scientist 15:8.
Alivisator AP, Blaser MJ, Brodie EL, et al. A unified initiative to harness Earth’s microbiomes. Science 350; 507-508.
Dubilier N, McFall-Ngai M, Zhao L. Create a global microbiome effort. Nature 2015; 526:631-634.
Relman DA, Schmidt TM, MacDermott RP, et al. Identification of the uncultured bacillus of Whipple’s disease. N Engl J Med 1992; 327:293-301.
Fredericks DN, Relman DA. 2001. Localization of Tropheryma whippelii rRNA in tissues from patients with Whipple’s disease. J Infect Dis 183:1229-1237.
Relman DA. 1999. The search for unrecognized pathogens. Science 284:1308-1310.
Lower R, Lower J, Kurth R. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA 93:5177-5184.
Hajjeh RA, Relman D, Cieslak PR, et al. 2002. Surveillance for unexplained deaths and critical illnesses due to possibly infectious causes, United States, 1995-1998. Emerg Infect Dis 8:145-153.
Gao SJ. Moore PS. 1996. Molecular approaches to the identification of unculturable infectious agent. Emerg Infect Dis 2:159-167.
Han YW, Shi W, Huang GT, et al. 2000. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun 68:3140-3146.
Prorok-Hamon M, Friswell MK, Alswied A, et al. 2014. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut 63:761-770.
Fujinami RS, von Herrath MG, Christen U, et al. 2006. Molecular Mimicry, Bystander Activation, or Viral Persistence: Infections and Autoimmune Disease. Clin Microbiol Rev 19:80-94. doi:10.1128/CMR.19.1.80-94.2006.
Hornig M. The role of microbes and autoimmunity in the pathogenesis of neuropsychiatric illness. Curr Opin Rheumatol 2013; 25:488-495.
Theiler M. Spontaneous encephalomyelitis of mice, a new virus disease. J Exp Med 1937; 65:705-719.
Yurkovetskiy LA, Pickard JM, Chervonsky AV. 2015. Microbiota and autoimmunity: exploring new avenues. Cell Host Microbe 17:548-552.